Welcome to animalgenomics@Mizzou. We envision using this site to disseminate information about our research program, which primarily focuses on beef and dairy cattle gene mapping although we have interests in other species (see projects). We are still developing the structure and content of the site so additional material will be posted in the near future. If you would like to be notified when we make major updates to the site please sign up for our newsletter to the left. We hope you find this information useful and welcome suggestions as to how to make it better.
The research in BDM lab is focused on developing machine learning and data mining methods to analyze big biomedical data for solving fundamental problems in biomedical sciences. Currently, we are developing bioinformatics algorithms and tools for protein structure and function prediction, 3D genomics, biological network modeling, and omics data analysis. The research is funded by both National Institutes of Health (NIH_ and National Science Foundation (NSF).
The Schultz-Appel Chemical Ecology Lab at the University of Missouri is directed by Jack Schultz and Heidi Appel of the Plant Sciences Division, College of Agriculture, Food and Natural Resources. Our lab addresses broad ecological and evolutionary questions in insect and plant science by using chemical, biochemical, and molecular methods.

Our lab develops powerful statistical and computational methods for the analysis of large and complex datasets, longitudinal data, survival data, and applied them to public health, medicine and social sciences, among others. In addition, we collaborate and consult with epidemiologists and clinicians on study design, data analysis and results interpretation.

The Decker Computational Genomics Group applies evolutionary biology to domesticated cattle, and other species as collaborations arise. We use the history of cattle populations to create genomic selection tools to improve cattle. Working at the intersection of two disciplines (bovine genomics and evolutionary biology) allows us to access large datasets, often produced for other research or commercial purposes. Our research program also has a transnational component in which we take basic research and transform it into decision support tools for farmers and ranchers.
The faculty and staff at the University of Missouri’s Department of Dermatology are dedicated to providing high quality dermatological care for our patients, exceptional educational experiences for our residents and students, and the ongoing pursuit of new knowledge and scholarship. We provide dermatology care for patients at University of Missouri Health Care Hospital and Clinics, the Harry S. Truman Memorial Veterans’ Hospital, and the Ellis Fischel Cancer Center. We also have been conducting teledermatology clinics in rural, underserved areas of Missouri for over 15 years.

The research focus of Digital Biology Laboratory (DBL) is Bioinformatics and Computational Biology. The lab works on development of novel computational methods, algorithms, software and information systems, as well as on broad applications of these tools and other informatics resources for various biological and medical problems. Research topics include application of deep learning in biological and medical data analysis and prediction, protein structure prediction, high-throughput biological data analysis, protein post-translation modification analysis, and computational biology studies of plants, cancers, and microbes. The lab has collaborated with dozens of research labs.

The iDAS Lab hosts trainees and researchers in computational science and informatics to design algorithms and develop methods for faster and more accurate predicative analytics and information search through large-scale information sources in biomedicine, engineering, and plant sciences with storytelling values for actionable decisions. Building upon rich research foundations, the lab has four active research directions in exploratory mining, Big Data search, visual knowledge retrievals, and block chains. All theoretical works developed in the iDAS lab are required to be tested on real-world data sets, such as HealthFacts® (60 million patients), National Impatient Sample (NIS), TCGA data, PDB, Simons Foundation Autism Research Initiative data, etc.

Dr. Meyers’s current research includes programs that emphasize bioinformatics and plant functional genomics. These programs include (1) analyses of small RNA, DNA methylation and the genomes of rice, Arabidopsis and other species using short-read DNA sequencing technologies, (2) development and implementation of novel informatics approaches for the storage, analysis, display, and public release of these data, (3) functional and evolutionary analyses of several gene families of interest, particularly miRNAs, phasiRNAs and the proteins involved in their biogenesis, as well as NB-LRR disease resistance genes.
The laboratory is directed by Dr. Henry T. Nguyen, Missouri Soybean Merchandising Council (MSMC) Endowed Professor of Genetics and Soybean Biotechnology. Research interests in the laboratory focus on the molecular genetics of plant stress tolerance and the application of genomics and genetic engineering technologies to soybean improvement methods.

By conducting and providing research in soybean genomics and biotechnology, we contribute to the genetic improvement of soybeans for food, human health, and industrial uses, while increasing the profitability of the U.S. soybean industry.
Visit the National Center for Soybean Biotechnology Website.

NextGen Biomedical Informatics (BMI), as part of the NextGen Precision Health Initiative and Data Science and Analytics Innovation Center (dSAIC), provides informatics expertise and capabilities to support translational and clinical researchers at all four University of Missouri System campuses, affiliated organizations and external partner organizations. NextGen BMI unites faculty and staff from the University of Missouri-Kansas City and the University of Missouri to leverage the capabilities of public-private partnerships such as the Tiger Institute for Health Innovation and our research infrastructure supported by NIH and PCORI funding. We are dedicated to developing and leveraging innovative informatics solutions and artificial intelligence and to advancing reproducible science by leading and participating in national research network collaborations.
Plant roots play a vital role in water and mineral acquisition, and are essential for plant growth and development. Under conditions of drought, roots can adapt to continue growth while at the same time producing and sending early warning signals to shoots which inhibit plant growth above ground. The plant root system, often referred to as “the hidden half,” has received much less attention compared to the shoot. We have formed a Plant Root Genomics Consortium dedicated to root genetics and physiology. The aim of this consortium is to develop an understanding of the molecular mechanisms used by plant roots to acquire water and minerals from the soil, to elucidate the role roots play in adaptation to drought conditions, and to transfer this knowledge to crop improvement through biotechnology.
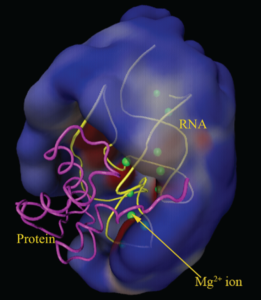
Dr. Chen’s lab has been developing computational methods for RNA structure and function and applications of the computational models to therapeutic designs. A main research method used in the lab is to design and implement knowledge-based database and to extract structure and function information from the database. Examples of the current research efforts include RNA 3D structure prediction based on rules derived from the known structure database, prediction of metal ion binding sites on nucleic acids through data training, sgRNA design and genome-wide off-target prediction for CRISPR-Cas9 gene editing system, and RNA-based drug design.

The TCBI lab conducts research in translational and cancer bioinformatics, imaging informatics, pathology informatics, computational epigenetics, and precision medicine informatics under the leadership of Dr. Dmitriy Shin. We draw from the experience of leading pathologists to study computational reasoning over molecular knowledge networks, function of long non-coding RNAs, molecular underpinnings of intra-tumor heterogeneity, brain damage imaging, diagnostics heuristics, pathway analysis, knowledge representation and knowledge complexity reduction, and many other topics. The lab is part of Pathology Informatics, a division of Pathology and Anatomical Sciences Department, and MU Informatics Institute.

At VIMAN Lab, we investigate novel methods to model, measure, manage and secure distributed computing applications that have real-time resource allocation needs and require high-speed networks. With broadband access and cloud computing becoming integral in our society, end-user applications (e.g., video-on-demand, videoconferencing, remote visualization, remote instrumentation, real-time data analytics) are increasingly becoming data-intensive, mobility-supported and network-dependent. Consequently, we are motivated by the challenges in studying theoretical foundations as well as developing tools for networks and cloud platforms. We strive for meeting the end-user Quality of Experience (QoE) expectations in applications (e.g., healthcare, advanced manufacturing, public safety, new media, bioinformatics).

In the ViGIR lab, we conduct research in areas such as: Computer Vision, Pattern Recognition and Robotics. We develop models to represent the world as perceived by cameras and other sensors and we devise algorithms that make use of such models to extract real time information from a sensory network in order to guide robots to perform various tasks. Our main goal is to build new Human-Robot Interfaces that can be used in robotic assistive technology, augmented reality, automation, tele-operation, etc….
Visit the Vision-Guided and Intelligent Robotics Lab (ViGIR) Website.

We are developing computational methods to calculate binding free energies for ligand-receptor complexes. The derived energy models are applied to protein-substrate interactions, protein-protein interactions, and structure-based drug design. We are also developing new docking algorithms to account for protein flexibility. Methods used in our laboratory include computer modeling, simulation and graphics display. Additional application includes modeling of structure-function relationship of membrane proteins.